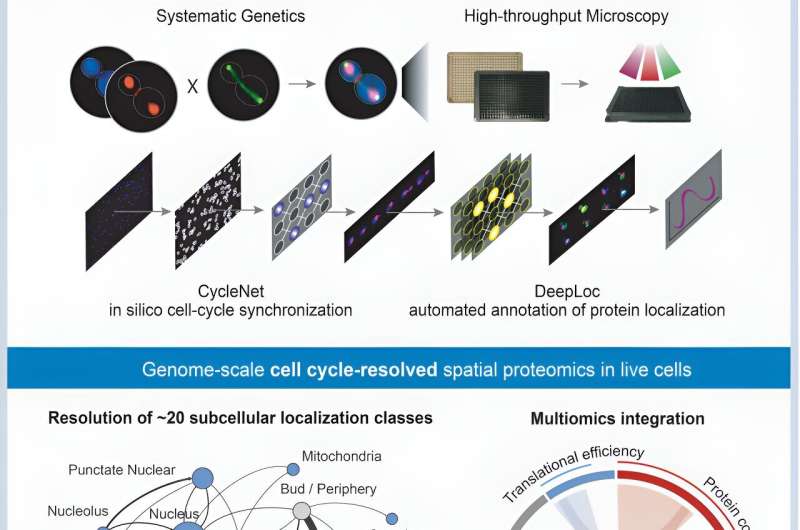
A study led by the University of Toronto has just seen the movement of proteins encoded by the yeast genome mapped throughout its cell cycle successfully. The process of monitoring all the proteins of an organism for the duration from the beginning of the cell cycle until the end represents the first instance of this kind. The research, which was recently published in a prestigious Cell journal, used the complex combination of deep learning technology and advanced microscopy to make this discovery possible.
Researchers tested two deep learning models namely DeepLoc and CycleNet to analyze yeast cells images of millions in real life. Through the implementation of this cutting-edge computational method, scientists produced the detailed map, pinpointing the exact places as well as the substitution process of proteins within the cell at each stage of the cell cycle.
The studies generate information about the delicate orchestration of proteins all through the cell cycle. It was their discovery that proteins are usually responsible for the cell cycle when they notice the expression patterns being regular when their levels in the cell keep fluctuating from time to time. On the other hand, proteins that move around the cell in a predictable manner are mostly involved in the physical enactment of the process.
Significance for biology and medicine
Understanding the cell cycle at a molecular level is indispensable as it is at the core of the cellular growth of all life organisms. The imbalance of proteins plays a crucial role in the regulation of the cell cycle, which, in turn, depending on the kind of disruption, could lead to serious diseases like cancer.
The research discovered hundreds of proteins that were located and concentrated at certain positions or levels during the different phases of the cell cycle. This brought forward the idea of multi-level regulation as a requirement for the cells to perhaps divide correctly. Through the use of fluorescence microscopy and machine learning algorithms, the researchers precisely profiled thousands of proteins throughout distinct cellular junctions, revealing previously unseen granularity in the proteins’ molecular choreography during cell division.
Future implications
The yeast cell is an irreplaceable model of eukaryotic biology which is determined by its specific features. The knowledge obtained from yeast cell studies can be used in understanding the human cell cycle, which can be of use in advancements in medicine and biotechnology.
The first author of the study, Dr. Athanasios Litsios, who is a postdoctoral fellow at the Donnelly Centre for Cellular and Biomolecular Research, said that the importance of yeast cells, as a model organism, in studying complicated biological processes at a genome-wide level cannot be overemphasized.
Finally, the study by the University of Toronto represents a major breakthrough in cellular biology. Through the use of cutting-edge technology and advanced computational approaches, the researchers have figured out the complex choreography of proteins inside yeast cells during the cell cycle. This historical study not only advances our understanding of basic biological processes but also opens a great potential for solving difficult medical problems in the future.
This article first appeared on the University of Toronto